12. Multivariate Normal Distribution#
Contents
12.1. Overview#
This lecture describes a workhorse in probability theory, statistics, and economics, namely, the multivariate normal distribution.
In this lecture, you will learn formulas for
the joint distribution of a random vector
marginal distributions for all subvectors of
conditional distributions for subvectors of
We will use the multivariate normal distribution to formulate some useful models:
a factor analytic model of an intelligence quotient, i.e., IQ
a factor analytic model of two independent inherent abilities, say, mathematical and verbal.
a more general factor analytic model
Principal Components Analysis (PCA) as an approximation to a factor analytic model
time series generated by linear stochastic difference equations
optimal linear filtering theory
12.2. The Multivariate Normal Distribution#
This lecture defines a Python class MultivariateNormal to be used
to generate marginal and conditional distributions associated
with a multivariate normal distribution.
For a multivariate normal distribution it is very convenient that
conditional expectations equal linear least squares projections
conditional distributions are characterized by multivariate linear regressions
We apply our Python class to some examples.
We use the following imports:
import matplotlib.pyplot as plt
plt.rcParams["figure.figsize"] = (11, 5) #set default figure size
import numpy as np
from numba import jit
import statsmodels.api as sm
Assume that an
This means that the probability density takes the form
where
The covariance matrix
@jit
def f(z, μ, Σ):
"""
The density function of multivariate normal distribution.
Parameters
---------------
z: ndarray(float, dim=2)
random vector, N by 1
μ: ndarray(float, dim=1 or 2)
the mean of z, N by 1
Σ: ndarray(float, dim=2)
the covarianece matrix of z, N by 1
"""
z = np.atleast_2d(z)
μ = np.atleast_2d(μ)
Σ = np.atleast_2d(Σ)
N = z.size
temp1 = np.linalg.det(Σ) ** (-1/2)
temp2 = np.exp(-.5 * (z - μ).T @ np.linalg.inv(Σ) @ (z - μ))
return (2 * np.pi) ** (-N/2) * temp1 * temp2
For some integer
where
Let
be corresponding partitions of
The marginal distribution of
multivariate normal with mean
The marginal distribution of
multivariate normal with mean
The distribution of
multivariate normal with mean
and covariance matrix
where
is an
The following class constructs a multivariate normal distribution instance with two methods.
a method
partitioncomputesa method
cond_distcomputes either the distribution of
class MultivariateNormal:
"""
Class of multivariate normal distribution.
Parameters
----------
μ: ndarray(float, dim=1)
the mean of z, N by 1
Σ: ndarray(float, dim=2)
the covarianece matrix of z, N by 1
Arguments
---------
μ, Σ:
see parameters
μs: list(ndarray(float, dim=1))
list of mean vectors μ1 and μ2 in order
Σs: list(list(ndarray(float, dim=2)))
2 dimensional list of covariance matrices
Σ11, Σ12, Σ21, Σ22 in order
βs: list(ndarray(float, dim=1))
list of regression coefficients β1 and β2 in order
"""
def __init__(self, μ, Σ):
"initialization"
self.μ = np.array(μ)
self.Σ = np.atleast_2d(Σ)
def partition(self, k):
"""
Given k, partition the random vector z into a size k vector z1
and a size N-k vector z2. Partition the mean vector μ into
μ1 and μ2, and the covariance matrix Σ into Σ11, Σ12, Σ21, Σ22
correspondingly. Compute the regression coefficients β1 and β2
using the partitioned arrays.
"""
μ = self.μ
Σ = self.Σ
self.μs = [μ[:k], μ[k:]]
self.Σs = [[Σ[:k, :k], Σ[:k, k:]],
[Σ[k:, :k], Σ[k:, k:]]]
self.βs = [self.Σs[0][1] @ np.linalg.inv(self.Σs[1][1]),
self.Σs[1][0] @ np.linalg.inv(self.Σs[0][0])]
def cond_dist(self, ind, z):
"""
Compute the conditional distribution of z1 given z2, or reversely.
Argument ind determines whether we compute the conditional
distribution of z1 (ind=0) or z2 (ind=1).
Returns
---------
μ_hat: ndarray(float, ndim=1)
The conditional mean of z1 or z2.
Σ_hat: ndarray(float, ndim=2)
The conditional covariance matrix of z1 or z2.
"""
β = self.βs[ind]
μs = self.μs
Σs = self.Σs
μ_hat = μs[ind] + β @ (z - μs[1-ind])
Σ_hat = Σs[ind][ind] - β @ Σs[1-ind][1-ind] @ β.T
return μ_hat, Σ_hat
Let’s put this code to work on a suite of examples.
We begin with a simple bivariate example; after that we’ll turn to a trivariate example.
We’ll compute population moments of some conditional distributions using
our MultivariateNormal class.
For fun we’ll also compute sample analogs of the associated population regressions by generating simulations and then computing linear least squares regressions.
We’ll compare those linear least squares regressions for the simulated data to their population counterparts.
12.3. Bivariate Example#
We start with a bivariate normal distribution pinned down by
μ = np.array([.5, 1.])
Σ = np.array([[1., .5], [.5 ,1.]])
# construction of the multivariate normal instance
multi_normal = MultivariateNormal(μ, Σ)
k = 1 # choose partition
# partition and compute regression coefficients
multi_normal.partition(k)
multi_normal.βs[0],multi_normal.βs[1]
(array([[0.5]]), array([[0.5]]))
Let’s illustrate the fact that you can regress anything on anything else.
We have computed everything we need to compute two regression lines, one of
We’ll represent these regressions as
and
where we have the population least squares orthogonality conditions
and
Let’s compute
beta = multi_normal.βs
a1 = μ[0] - beta[0]*μ[1]
b1 = beta[0]
a2 = μ[1] - beta[1]*μ[0]
b2 = beta[1]
Let’s print out the intercepts and slopes.
For the regression of
print ("a1 = ", a1)
print ("b1 = ", b1)
a1 = [[0.]]
b1 = [[0.5]]
For the regression of
print ("a2 = ", a2)
print ("b2 = ", b2)
a2 = [[0.75]]
b2 = [[0.5]]
Now let’s plot the two regression lines and stare at them.
z2 = np.linspace(-4,4,100)
a1 = np.squeeze(a1)
b1 = np.squeeze(b1)
a2 = np.squeeze(a2)
b2 = np.squeeze(b2)
z1 = b1*z2 + a1
z1h = z2/b2 - a2/b2
fig = plt.figure(figsize=(12,12))
ax = fig.add_subplot(1, 1, 1)
ax.set(xlim=(-4, 4), ylim=(-4, 4))
ax.spines['left'].set_position('center')
ax.spines['bottom'].set_position('zero')
ax.spines['right'].set_color('none')
ax.spines['top'].set_color('none')
ax.xaxis.set_ticks_position('bottom')
ax.yaxis.set_ticks_position('left')
plt.ylabel('$z_1$', loc = 'top')
plt.xlabel('$z_2$,', loc = 'right')
plt.title('two regressions')
plt.plot(z2,z1, 'r', label = "$z_1$ on $z_2$")
plt.plot(z2,z1h, 'b', label = "$z_2$ on $z_1$")
plt.legend()
plt.show()
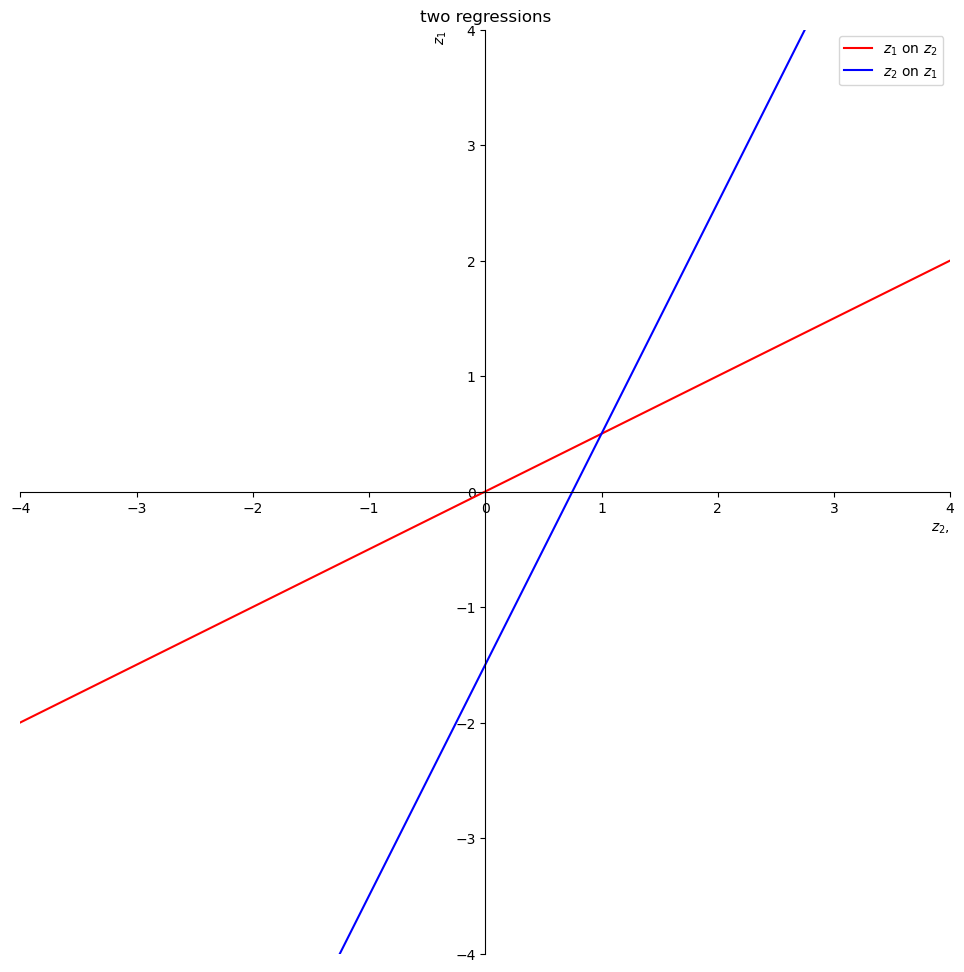
The red line is the expectation of
The intercept and slope of the red line are
print("a1 = ", a1)
print("b1 = ", b1)
a1 = 0.0
b1 = 0.5
The blue line is the expectation of
The intercept and slope of the blue line are
print("-a2/b2 = ", - a2/b2)
print("1/b2 = ", 1/b2)
-a2/b2 = -1.5
1/b2 = 2.0
We can use these regression lines or our code to compute conditional expectations.
Let’s compute the mean and variance of the distribution of
After that we’ll reverse what are on the left and right sides of the regression.
# compute the cond. dist. of z1
ind = 1
z1 = np.array([5.]) # given z1
μ2_hat, Σ2_hat = multi_normal.cond_dist(ind, z1)
print('μ2_hat, Σ2_hat = ', μ2_hat, Σ2_hat)
μ2_hat, Σ2_hat = [3.25] [[0.75]]
Now let’s compute the mean and variance of the distribution of
# compute the cond. dist. of z1
ind = 0
z2 = np.array([5.]) # given z2
μ1_hat, Σ1_hat = multi_normal.cond_dist(ind, z2)
print('μ1_hat, Σ1_hat = ', μ1_hat, Σ1_hat)
μ1_hat, Σ1_hat = [2.5] [[0.75]]
Let’s compare the preceding population mean and variance with outcomes
from drawing a large sample and then regressing
We know that
which can be arranged to
We anticipate that for larger and larger sample sizes, estimated OLS
coefficients will converge to
n = 1_000_000 # sample size
# simulate multivariate normal random vectors
data = np.random.multivariate_normal(μ, Σ, size=n)
z1_data = data[:, 0]
z2_data = data[:, 1]
# OLS regression
μ1, μ2 = multi_normal.μs
results = sm.OLS(z1_data - μ1, z2_data - μ2).fit()
Let’s compare the preceding population
multi_normal.βs[0], results.params
(array([[0.5]]), array([0.50023899]))
Let’s compare our population
Σ1_hat, results.resid @ results.resid.T / (n - 1)
(array([[0.75]]), 0.7501698706292744)
Lastly, let’s compute the estimate of
μ1_hat, results.predict(z2 - μ2) + μ1
(array([2.5]), array([2.50095595]))
Thus, in each case, for our very large sample size, the sample analogues closely approximate their population counterparts.
A Law of Large Numbers explains why sample analogues approximate population objects.
12.4. Trivariate Example#
Let’s apply our code to a trivariate example.
We’ll specify the mean vector and the covariance matrix as follows.
μ = np.random.random(3)
C = np.random.random((3, 3))
Σ = C @ C.T # positive semi-definite
multi_normal = MultivariateNormal(μ, Σ)
μ, Σ
(array([0.70655948, 0.41884944, 0.57227581]),
array([[0.28459369, 0.26256164, 0.38978302],
[0.26256164, 0.51197677, 0.36294358],
[0.38978302, 0.36294358, 1.53707536]]))
k = 1
multi_normal.partition(k)
Let’s compute the distribution of
ind = 0
z2 = np.array([2., 5.])
μ1_hat, Σ1_hat = multi_normal.cond_dist(ind, z2)
n = 1_000_000
data = np.random.multivariate_normal(μ, Σ, size=n)
z1_data = data[:, :k]
z2_data = data[:, k:]
μ1, μ2 = multi_normal.μs
results = sm.OLS(z1_data - μ1, z2_data - μ2).fit()
As above, we compare population and sample regression coefficients, the conditional covariance matrix, and the conditional mean vector in that order.
multi_normal.βs[0], results.params
(array([[0.40003092, 0.15912971]]), array([0.40048621, 0.1585142 ]))
Σ1_hat, results.resid @ results.resid.T / (n - 1)
(array([[0.11753485]]), 0.1178189244754668)
μ1_hat, results.predict(z2 - μ2) + μ1
(array([2.04365108]), array([2.04164564]))
Once again, sample analogues do a good job of approximating their populations counterparts.
12.5. One Dimensional Intelligence (IQ)#
Let’s move closer to a real-life example, namely, inferring a one-dimensional measure of intelligence called IQ from a list of test scores.
The
The distribution of IQ’s for a cross-section of people is a normal random variable described by
We assume that the noises
We also assume that
The following system describes the
or equivalently,
where
Let’s define a Python function that constructs the mean
As arguments, the function takes the number of tests
def construct_moments_IQ(n, μθ, σθ, σy):
μ_IQ = np.full(n+1, μθ)
D_IQ = np.zeros((n+1, n+1))
D_IQ[range(n), range(n)] = σy
D_IQ[:, n] = σθ
Σ_IQ = D_IQ @ D_IQ.T
return μ_IQ, Σ_IQ, D_IQ
Now let’s consider a specific instance of this model.
Assume we have recorded
We can compute the mean vector and covariance matrix of construct_moments_IQ function as follows.
n = 50
μθ, σθ, σy = 100., 10., 10.
μ_IQ, Σ_IQ, D_IQ = construct_moments_IQ(n, μθ, σθ, σy)
μ_IQ, Σ_IQ, D_IQ
(array([100., 100., 100., 100., 100., 100., 100., 100., 100., 100., 100.,
100., 100., 100., 100., 100., 100., 100., 100., 100., 100., 100.,
100., 100., 100., 100., 100., 100., 100., 100., 100., 100., 100.,
100., 100., 100., 100., 100., 100., 100., 100., 100., 100., 100.,
100., 100., 100., 100., 100., 100., 100.]),
array([[200., 100., 100., ..., 100., 100., 100.],
[100., 200., 100., ..., 100., 100., 100.],
[100., 100., 200., ..., 100., 100., 100.],
...,
[100., 100., 100., ..., 200., 100., 100.],
[100., 100., 100., ..., 100., 200., 100.],
[100., 100., 100., ..., 100., 100., 100.]]),
array([[10., 0., 0., ..., 0., 0., 10.],
[ 0., 10., 0., ..., 0., 0., 10.],
[ 0., 0., 10., ..., 0., 0., 10.],
...,
[ 0., 0., 0., ..., 10., 0., 10.],
[ 0., 0., 0., ..., 0., 10., 10.],
[ 0., 0., 0., ..., 0., 0., 10.]]))
We can now use our MultivariateNormal class to construct an
instance, then partition the mean vector and covariance matrix as we
wish.
We want to regress IQ, the random variable
We choose k=n so that
multi_normal_IQ = MultivariateNormal(μ_IQ, Σ_IQ)
k = n
multi_normal_IQ.partition(k)
Using the generator multivariate_normal, we can make one draw of the
random vector from our distribution and then compute the distribution of
Let’s do that and then print out some pertinent quantities.
x = np.random.multivariate_normal(μ_IQ, Σ_IQ)
y = x[:-1] # test scores
θ = x[-1] # IQ
# the true value
θ
98.40800901560205
The method cond_dist takes test scores
In the following code, ind sets the variables on the right side of the regression.
Given the way we have defined the vector ind=1 in order to make
ind = 1
multi_normal_IQ.cond_dist(ind, y)
(array([97.35083863]), array([[1.96078431]]))
The first number is the conditional mean
How do additional test scores affect our inferences?
To shed light on this, we compute a sequence of conditional
distributions of
We’ll make a pretty graph showing how our judgment of the person’s IQ change as more test results come in.
# array for containing moments
μθ_hat_arr = np.empty(n)
Σθ_hat_arr = np.empty(n)
# loop over number of test scores
for i in range(1, n+1):
# construction of multivariate normal distribution instance
μ_IQ_i, Σ_IQ_i, D_IQ_i = construct_moments_IQ(i, μθ, σθ, σy)
multi_normal_IQ_i = MultivariateNormal(μ_IQ_i, Σ_IQ_i)
# partition and compute conditional distribution
multi_normal_IQ_i.partition(i)
scores_i = y[:i]
μθ_hat_i, Σθ_hat_i = multi_normal_IQ_i.cond_dist(1, scores_i)
# store the results
μθ_hat_arr[i-1] = μθ_hat_i[0]
Σθ_hat_arr[i-1] = Σθ_hat_i[0, 0]
# transform variance to standard deviation
σθ_hat_arr = np.sqrt(Σθ_hat_arr)
μθ_hat_lower = μθ_hat_arr - 1.96 * σθ_hat_arr
μθ_hat_higher = μθ_hat_arr + 1.96 * σθ_hat_arr
plt.hlines(θ, 1, n+1, ls='--', label='true $θ$')
plt.plot(range(1, n+1), μθ_hat_arr, color='b', label=r'$\hat{μ}_{θ}$')
plt.plot(range(1, n+1), μθ_hat_lower, color='b', ls='--')
plt.plot(range(1, n+1), μθ_hat_higher, color='b', ls='--')
plt.fill_between(range(1, n+1), μθ_hat_lower, μθ_hat_higher,
color='b', alpha=0.2, label='95%')
plt.xlabel('number of test scores')
plt.ylabel('$\hat{θ}$')
plt.legend()
plt.show()
<>:12: SyntaxWarning: invalid escape sequence '\h'
<>:12: SyntaxWarning: invalid escape sequence '\h'
/tmp/ipykernel_8076/3550752491.py:12: SyntaxWarning: invalid escape sequence '\h'
plt.ylabel('$\hat{θ}$')
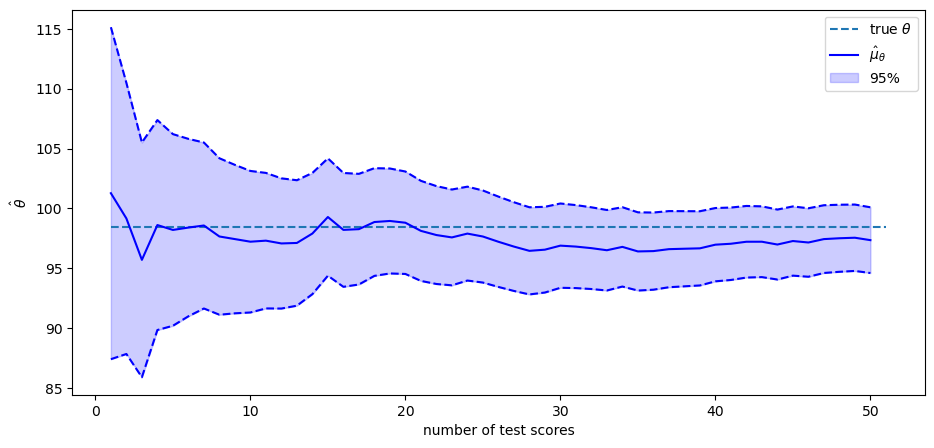
The solid blue line in the plot above shows
The blue area shows the span that comes from adding or subtracting
Therefore,
The value of the random
As more and more test scores come in, our estimate of the person’s
By staring at the changes in the conditional distributions, we see that
adding more test scores makes
Thus, each
If we were to drive the number of tests
12.6. Information as Surprise#
By using a different representation, let’s look at things from a different perspective.
We can represent the random vector
where
and
It follows that
Let
We can compute
This formula confirms that the orthonormal vector
We can say that
Let
Then we can write
The mutual orthogonality of the
Thus, relative to what is known from tests
Here new information means surprise or what could not be predicted from earlier information.
Formula (12.1) also provides us with an enlightening way to express conditional means and conditional variances that we computed earlier.
In particular,
and
C = np.linalg.cholesky(Σ_IQ)
G = np.linalg.inv(C)
ε = G @ (x - μθ)
cε = C[n, :] * ε
# compute the sequence of μθ and Σθ conditional on y1, y2, ..., yk
μθ_hat_arr_C = np.array([np.sum(cε[:k+1]) for k in range(n)]) + μθ
Σθ_hat_arr_C = np.array([np.sum(C[n, i+1:n+1] ** 2) for i in range(n)])
To confirm that these formulas give the same answers that we computed
earlier, we can compare the means and variances of MultivariateNormal built on
our original representation of conditional distributions for
multivariate normal distributions.
# conditional mean
np.max(np.abs(μθ_hat_arr - μθ_hat_arr_C)) < 1e-10
True
# conditional variance
np.max(np.abs(Σθ_hat_arr - Σθ_hat_arr_C)) < 1e-10
True
12.7. Cholesky Factor Magic#
Evidently, the Cholesky factorizations automatically computes the
population regression coefficients and associated statistics
that are produced by our MultivariateNormal class.
The Cholesky factorization computes these things recursively.
Indeed, in formula (12.1),
the random variable
the coefficient
12.8. Math and Verbal Intelligence#
We can alter the preceding example to be more realistic.
There is ample evidence that IQ is not a scalar.
Some people are good in math skills but poor in language skills.
Other people are good in language skills but poor in math skills.
So now we shall assume that there are two dimensions of IQ,
These determine average performances in math and language tests, respectively.
We observe math scores
When
where
We construct a Python function construct_moments_IQ2d to construct
the mean vector and covariance matrix of the joint normal distribution.
def construct_moments_IQ2d(n, μθ, σθ, μη, ση, σy):
μ_IQ2d = np.empty(2*(n+1))
μ_IQ2d[:n] = μθ
μ_IQ2d[2*n] = μθ
μ_IQ2d[n:2*n] = μη
μ_IQ2d[2*n+1] = μη
D_IQ2d = np.zeros((2*(n+1), 2*(n+1)))
D_IQ2d[range(2*n), range(2*n)] = σy
D_IQ2d[:n, 2*n] = σθ
D_IQ2d[2*n, 2*n] = σθ
D_IQ2d[n:2*n, 2*n+1] = ση
D_IQ2d[2*n+1, 2*n+1] = ση
Σ_IQ2d = D_IQ2d @ D_IQ2d.T
return μ_IQ2d, Σ_IQ2d, D_IQ2d
Let’s put the function to work.
n = 2
# mean and variance of θ, η, and y
μθ, σθ, μη, ση, σy = 100., 10., 100., 10, 10
μ_IQ2d, Σ_IQ2d, D_IQ2d = construct_moments_IQ2d(n, μθ, σθ, μη, ση, σy)
μ_IQ2d, Σ_IQ2d, D_IQ2d
(array([100., 100., 100., 100., 100., 100.]),
array([[200., 100., 0., 0., 100., 0.],
[100., 200., 0., 0., 100., 0.],
[ 0., 0., 200., 100., 0., 100.],
[ 0., 0., 100., 200., 0., 100.],
[100., 100., 0., 0., 100., 0.],
[ 0., 0., 100., 100., 0., 100.]]),
array([[10., 0., 0., 0., 10., 0.],
[ 0., 10., 0., 0., 10., 0.],
[ 0., 0., 10., 0., 0., 10.],
[ 0., 0., 0., 10., 0., 10.],
[ 0., 0., 0., 0., 10., 0.],
[ 0., 0., 0., 0., 0., 10.]]))
# take one draw
x = np.random.multivariate_normal(μ_IQ2d, Σ_IQ2d)
y1 = x[:n]
y2 = x[n:2*n]
θ = x[2*n]
η = x[2*n+1]
# the true values
θ, η
(98.25677374913887, 101.41570209166122)
We first compute the joint normal distribution of
multi_normal_IQ2d = MultivariateNormal(μ_IQ2d, Σ_IQ2d)
k = 2*n # the length of data vector
multi_normal_IQ2d.partition(k)
multi_normal_IQ2d.cond_dist(1, [*y1, *y2])
(array([96.11974166, 97.0216911 ]),
array([[33.33333333, 0. ],
[ 0. , 33.33333333]]))
Now let’s compute distributions of
It will be fun to compare outcomes with the help of an auxiliary function
cond_dist_IQ2d that we now construct.
def cond_dist_IQ2d(μ, Σ, data):
n = len(μ)
multi_normal = MultivariateNormal(μ, Σ)
multi_normal.partition(n-1)
μ_hat, Σ_hat = multi_normal.cond_dist(1, data)
return μ_hat, Σ_hat
Let’s see how things work for an example.
for indices, IQ, conditions in [([*range(2*n), 2*n], 'θ', 'y1, y2, y3, y4'),
([*range(n), 2*n], 'θ', 'y1, y2'),
([*range(n, 2*n), 2*n], 'θ', 'y3, y4'),
([*range(2*n), 2*n+1], 'η', 'y1, y2, y3, y4'),
([*range(n), 2*n+1], 'η', 'y1, y2'),
([*range(n, 2*n), 2*n+1], 'η', 'y3, y4')]:
μ_hat, Σ_hat = cond_dist_IQ2d(μ_IQ2d[indices], Σ_IQ2d[indices][:, indices], x[indices[:-1]])
print(f'The mean and variance of {IQ} conditional on {conditions: <15} are ' +
f'{μ_hat[0]:1.2f} and {Σ_hat[0, 0]:1.2f} respectively')
The mean and variance of θ conditional on y1, y2, y3, y4 are 96.12 and 33.33 respectively
The mean and variance of θ conditional on y1, y2 are 96.12 and 33.33 respectively
The mean and variance of θ conditional on y3, y4 are 100.00 and 100.00 respectively
The mean and variance of η conditional on y1, y2, y3, y4 are 97.02 and 33.33 respectively
The mean and variance of η conditional on y1, y2 are 100.00 and 100.00 respectively
The mean and variance of η conditional on y3, y4 are 97.02 and 33.33 respectively
Evidently, math tests provide no information about
12.9. Univariate Time Series Analysis#
We can use the multivariate normal distribution and a little matrix algebra to present foundations of univariate linear time series analysis.
Let
Consider the following model:
We can compute the moments of
Given some
and the covariance matrix
Similarly, we can define
and therefore
where
Consequently, the covariance matrix of
By stacking
and
Thus, the stacked sequences
# as an example, consider the case where T = 3
T = 3
# variance of the initial distribution x_0
σ0 = 1.
# parameters of the equation system
a = .9
b = 1.
c = 1.0
d = .05
# construct the covariance matrix of X
Σx = np.empty((T+1, T+1))
Σx[0, 0] = σ0 ** 2
for i in range(T):
Σx[i, i+1:] = Σx[i, i] * a ** np.arange(1, T+1-i)
Σx[i+1:, i] = Σx[i, i+1:]
Σx[i+1, i+1] = a ** 2 * Σx[i, i] + b ** 2
Σx
array([[1. , 0.9 , 0.81 , 0.729 ],
[0.9 , 1.81 , 1.629 , 1.4661 ],
[0.81 , 1.629 , 2.4661 , 2.21949 ],
[0.729 , 1.4661 , 2.21949 , 2.997541]])
# construct the covariance matrix of Y
C = np.eye(T+1) * c
D = np.eye(T+1) * d
Σy = C @ Σx @ C.T + D @ D.T
# construct the covariance matrix of Z
Σz = np.empty((2*(T+1), 2*(T+1)))
Σz[:T+1, :T+1] = Σx
Σz[:T+1, T+1:] = Σx @ C.T
Σz[T+1:, :T+1] = C @ Σx
Σz[T+1:, T+1:] = Σy
Σz
array([[1. , 0.9 , 0.81 , 0.729 , 1. , 0.9 ,
0.81 , 0.729 ],
[0.9 , 1.81 , 1.629 , 1.4661 , 0.9 , 1.81 ,
1.629 , 1.4661 ],
[0.81 , 1.629 , 2.4661 , 2.21949 , 0.81 , 1.629 ,
2.4661 , 2.21949 ],
[0.729 , 1.4661 , 2.21949 , 2.997541, 0.729 , 1.4661 ,
2.21949 , 2.997541],
[1. , 0.9 , 0.81 , 0.729 , 1.0025 , 0.9 ,
0.81 , 0.729 ],
[0.9 , 1.81 , 1.629 , 1.4661 , 0.9 , 1.8125 ,
1.629 , 1.4661 ],
[0.81 , 1.629 , 2.4661 , 2.21949 , 0.81 , 1.629 ,
2.4686 , 2.21949 ],
[0.729 , 1.4661 , 2.21949 , 2.997541, 0.729 , 1.4661 ,
2.21949 , 3.000041]])
# construct the mean vector of Z
μz = np.zeros(2*(T+1))
The following Python code lets us sample random vectors
This is going to be very useful for doing the conditioning to be used in the fun exercises below.
z = np.random.multivariate_normal(μz, Σz)
x = z[:T+1]
y = z[T+1:]
12.9.1. Smoothing Example#
This is an instance of a classic smoothing calculation whose purpose
is to compute
An interpretation of this example is
# construct a MultivariateNormal instance
multi_normal_ex1 = MultivariateNormal(μz, Σz)
x = z[:T+1]
y = z[T+1:]
# partition Z into X and Y
multi_normal_ex1.partition(T+1)
# compute the conditional mean and covariance matrix of X given Y=y
print("X = ", x)
print("Y = ", y)
print(" E [ X | Y] = ", )
multi_normal_ex1.cond_dist(0, y)
X = [-1.72795143 -0.7321461 -3.84659701 -4.2759139 ]
Y = [-1.77368314 -0.72845598 -3.84781329 -4.30029643]
E [ X | Y] =
(array([-1.76734581, -0.7377382 , -3.84176018, -4.2981949 ]),
array([[2.48875094e-03, 5.57449314e-06, 1.24861729e-08, 2.80235835e-11],
[5.57449314e-06, 2.48876343e-03, 5.57452116e-06, 1.25113941e-08],
[1.24861729e-08, 5.57452116e-06, 2.48876346e-03, 5.58575339e-06],
[2.80235835e-11, 1.25113941e-08, 5.58575339e-06, 2.49377812e-03]]))
12.9.2. Filtering Exercise#
Compute
To do so, we need to first construct the mean vector and the covariance
matrix of the subvector
For example, let’s say that we want the conditional distribution of
t = 3
# mean of the subvector
sub_μz = np.zeros(t+1)
# covariance matrix of the subvector
sub_Σz = np.empty((t+1, t+1))
sub_Σz[0, 0] = Σz[t, t] # x_t
sub_Σz[0, 1:] = Σz[t, T+1:T+t+1]
sub_Σz[1:, 0] = Σz[T+1:T+t+1, t]
sub_Σz[1:, 1:] = Σz[T+1:T+t+1, T+1:T+t+1]
sub_Σz
array([[2.997541, 0.729 , 1.4661 , 2.21949 ],
[0.729 , 1.0025 , 0.9 , 0.81 ],
[1.4661 , 0.9 , 1.8125 , 1.629 ],
[2.21949 , 0.81 , 1.629 , 2.4686 ]])
multi_normal_ex2 = MultivariateNormal(sub_μz, sub_Σz)
multi_normal_ex2.partition(1)
sub_y = y[:t]
multi_normal_ex2.cond_dist(0, sub_y)
(array([-3.45588616]), array([[1.00201996]]))
12.9.3. Prediction Exercise#
Compute
As what we did in exercise 2, we will construct the mean vector and
covariance matrix of the subvector
For example, we take a case in which
t = 3
j = 2
sub_μz = np.zeros(t-j+2)
sub_Σz = np.empty((t-j+2, t-j+2))
sub_Σz[0, 0] = Σz[T+t+1, T+t+1]
sub_Σz[0, 1:] = Σz[T+t+1, T+1:T+t-j+2]
sub_Σz[1:, 0] = Σz[T+1:T+t-j+2, T+t+1]
sub_Σz[1:, 1:] = Σz[T+1:T+t-j+2, T+1:T+t-j+2]
sub_Σz
array([[3.000041, 0.729 , 1.4661 ],
[0.729 , 1.0025 , 0.9 ],
[1.4661 , 0.9 , 1.8125 ]])
multi_normal_ex3 = MultivariateNormal(sub_μz, sub_Σz)
multi_normal_ex3.partition(1)
sub_y = y[:t-j+1]
multi_normal_ex3.cond_dist(0, sub_y)
(array([-0.59179082]), array([[1.81413617]]))
12.9.4. Constructing a Wold Representation#
Now we’ll apply Cholesky decomposition to decompose
Then we can represent
H = np.linalg.cholesky(Σy)
H
array([[1.00124922, 0. , 0. , 0. ],
[0.8988771 , 1.00225743, 0. , 0. ],
[0.80898939, 0.89978675, 1.00225743, 0. ],
[0.72809046, 0.80980808, 0.89978676, 1.00225743]])
ε = np.linalg.inv(H) @ y
ε
array([-1.77147019, 0.86193226, -3.18308234, -0.84250836])
y
array([-1.77368314, -0.72845598, -3.84781329, -4.30029643])
This example is an instance of what is known as a Wold representation in time series analysis.
12.10. Stochastic Difference Equation#
Consider the stochastic second-order linear difference equation
where
It can be written as a stacked system
We can compute
We have
where
# set parameters
T = 80
T = 160
# coefficients of the second order difference equation
𝛼0 = 10
𝛼1 = 1.53
𝛼2 = -.9
# variance of u
σu = 1.
σu = 10.
# distribution of y_{-1} and y_{0}
μy_tilde = np.array([1., 0.5])
Σy_tilde = np.array([[2., 1.], [1., 0.5]])
# construct A and A^{\prime}
A = np.zeros((T, T))
for i in range(T):
A[i, i] = 1
if i-1 >= 0:
A[i, i-1] = -𝛼1
if i-2 >= 0:
A[i, i-2] = -𝛼2
A_inv = np.linalg.inv(A)
# compute the mean vectors of b and y
μb = np.full(T, 𝛼0)
μb[0] += 𝛼1 * μy_tilde[1] + 𝛼2 * μy_tilde[0]
μb[1] += 𝛼2 * μy_tilde[1]
μy = A_inv @ μb
# compute the covariance matrices of b and y
Σu = np.eye(T) * σu ** 2
Σb = np.zeros((T, T))
C = np.array([[𝛼2, 𝛼1], [0, 𝛼2]])
Σb[:2, :2] = C @ Σy_tilde @ C.T
Σy = A_inv @ (Σb + Σu) @ A_inv.T
12.11. Application to Stock Price Model#
Let
Form
we have
β = .96
# construct B
B = np.zeros((T, T))
for i in range(T):
B[i, i:] = β ** np.arange(0, T-i)
Denote
Thus,
D = np.vstack([np.eye(T), B])
μz = D @ μy
Σz = D @ Σy @ D.T
We can simulate paths of MultivariateNormal class.
z = np.random.multivariate_normal(μz, Σz)
y, p = z[:T], z[T:]
cond_Ep = np.empty(T-1)
sub_μ = np.empty(3)
sub_Σ = np.empty((3, 3))
for t in range(2, T+1):
sub_μ[:] = μz[[t-2, t-1, T-1+t]]
sub_Σ[:, :] = Σz[[t-2, t-1, T-1+t], :][:, [t-2, t-1, T-1+t]]
multi_normal = MultivariateNormal(sub_μ, sub_Σ)
multi_normal.partition(2)
cond_Ep[t-2] = multi_normal.cond_dist(1, y[t-2:t])[0][0]
plt.plot(range(1, T), y[1:], label='$y_{t}$')
plt.plot(range(1, T), y[:-1], label='$y_{t-1}$')
plt.plot(range(1, T), p[1:], label='$p_{t}$')
plt.plot(range(1, T), cond_Ep, label='$Ep_{t}|y_{t}, y_{t-1}$')
plt.xlabel('t')
plt.legend(loc=1)
plt.show()
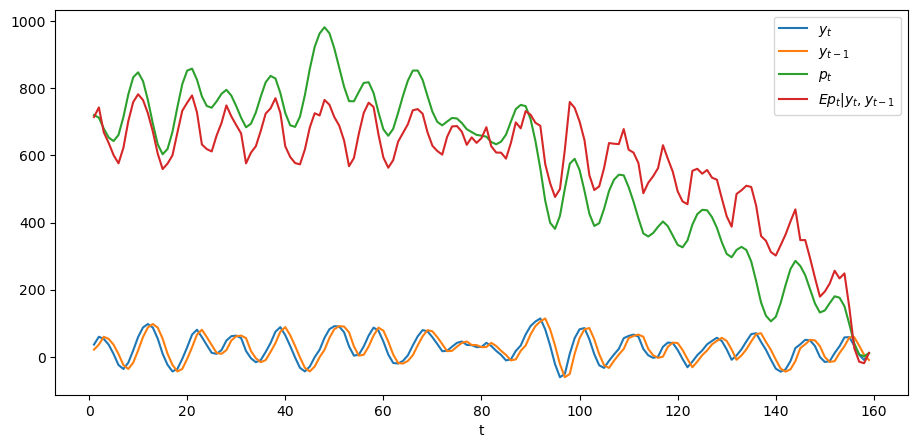
In the above graph, the green line is what the price of the stock would
be if people had perfect foresight about the path of dividends while the
green line is the conditional expectation
12.12. Filtering Foundations#
Assume that
where
We consider the problem of someone who
observes
does not observe
knows
wants to infer
Therefore, the person wants to construct the probability distribution of
The joint distribution of
By applying an appropriate instance of the above formulas for the mean vector
We can express our finding that the probability distribution of
where
12.12.1. Step toward dynamics#
Now suppose that we are in a time series setting and that we have the one-step state transition equation
where
Using equation (12.2), we can also represent
It follows that
and that the corresponding conditional covariance matrix
or
We can write the mean of
or
where
12.12.2. Dynamic version#
Suppose now that for
where as before
The logic and
formulas that we applied above imply that the probability distribution
of
where
If we shift the first equation forward one period and then substitute the expression for
This is a matrix Riccati difference equation that is closely related to another matrix Riccati difference equation that appears in a quantecon lecture on the basics of linear quadratic control theory.
That equation has the form
Stare at the two preceding equations for a moment or two, the first being a matrix difference equation for a conditional covariance matrix, the second being a matrix difference equation in the matrix appearing in a quadratic form for an intertemporal cost of value function.
Although the two equations are not identical, they display striking family resemblences.
the first equation tells dynamics that work forward in time
the second equation tells dynamics that work backward in time
while many of the terms are similar, one equation seems to apply matrix transformations to some matrices that play similar roles in the other equation
The family resemblences of these two equations reflects a transcendent duality that prevails between control theory and filtering theory.
12.12.3. An example#
We can use the Python class MultivariateNormal to construct examples.
Here is an example for a single period problem at time
G = np.array([[1., 3.]])
R = np.array([[1.]])
x0_hat = np.array([0., 1.])
Σ0 = np.array([[1., .5], [.3, 2.]])
μ = np.hstack([x0_hat, G @ x0_hat])
Σ = np.block([[Σ0, Σ0 @ G.T], [G @ Σ0, G @ Σ0 @ G.T + R]])
# construction of the multivariate normal instance
multi_normal = MultivariateNormal(μ, Σ)
multi_normal.partition(2)
# the observation of y
y0 = 2.3
# conditional distribution of x0
μ1_hat, Σ11 = multi_normal.cond_dist(0, y0)
μ1_hat, Σ11
(array([-0.078125, 0.803125]),
array([[ 0.72098214, -0.203125 ],
[-0.403125 , 0.228125 ]]))
A = np.array([[0.5, 0.2], [-0.1, 0.3]])
C = np.array([[2.], [1.]])
# conditional distribution of x1
x1_cond = A @ μ1_hat
Σ1_cond = C @ C.T + A @ Σ11 @ A.T
x1_cond, Σ1_cond
(array([0.1215625, 0.24875 ]),
array([[4.12874554, 1.95523214],
[1.92123214, 1.04592857]]))
12.12.4. Code for Iterating#
Here is code for solving a dynamic filtering problem by iterating on our equations, followed by an example.
def iterate(x0_hat, Σ0, A, C, G, R, y_seq):
p, n = G.shape
T = len(y_seq)
x_hat_seq = np.empty((T+1, n))
Σ_hat_seq = np.empty((T+1, n, n))
x_hat_seq[0] = x0_hat
Σ_hat_seq[0] = Σ0
for t in range(T):
xt_hat = x_hat_seq[t]
Σt = Σ_hat_seq[t]
μ = np.hstack([xt_hat, G @ xt_hat])
Σ = np.block([[Σt, Σt @ G.T], [G @ Σt, G @ Σt @ G.T + R]])
# filtering
multi_normal = MultivariateNormal(μ, Σ)
multi_normal.partition(n)
x_tilde, Σ_tilde = multi_normal.cond_dist(0, y_seq[t])
# forecasting
x_hat_seq[t+1] = A @ x_tilde
Σ_hat_seq[t+1] = C @ C.T + A @ Σ_tilde @ A.T
return x_hat_seq, Σ_hat_seq
iterate(x0_hat, Σ0, A, C, G, R, [2.3, 1.2, 3.2])
(array([[0. , 1. ],
[0.1215625 , 0.24875 ],
[0.18680212, 0.06904689],
[0.75576875, 0.05558463]]),
array([[[1. , 0.5 ],
[0.3 , 2. ]],
[[4.12874554, 1.95523214],
[1.92123214, 1.04592857]],
[[4.08198663, 1.99218488],
[1.98640488, 1.00886423]],
[[4.06457628, 2.00041999],
[1.99943739, 1.00275526]]]))
The iterative algorithm just described is a version of the celebrated Kalman filter.
We describe the Kalman filter and some applications of it in A First Look at the Kalman Filter
12.13. Classic Factor Analysis Model#
The factor analysis model widely used in psychology and other fields can be represented as
where
It is presumed that
This implies that
Thus, the covariance matrix
This means that all covariances among the
Form
the covariance matrix of the expanded random vector
In the following, we first construct the mean vector and the covariance
matrix for the case where
N = 10
k = 2
We set the coefficient matrix
where the first half of the first column of
Λ = np.zeros((N, k))
Λ[:N//2, 0] = 1
Λ[N//2:, 1] = 1
σu = .5
D = np.eye(N) * σu ** 2
# compute Σy
Σy = Λ @ Λ.T + D
We can now construct the mean vector and the covariance matrix for
μz = np.zeros(k+N)
Σz = np.empty((k+N, k+N))
Σz[:k, :k] = np.eye(k)
Σz[:k, k:] = Λ.T
Σz[k:, :k] = Λ
Σz[k:, k:] = Σy
z = np.random.multivariate_normal(μz, Σz)
f = z[:k]
y = z[k:]
multi_normal_factor = MultivariateNormal(μz, Σz)
multi_normal_factor.partition(k)
Let’s compute the conditional distribution of the hidden factor
multi_normal_factor.cond_dist(0, y)
(array([-1.14557316, 0.38088811]),
array([[0.04761905, 0. ],
[0. , 0.04761905]]))
We can verify that the conditional mean
B = Λ.T @ np.linalg.inv(Σy)
B @ y
array([-1.14557316, 0.38088811])
Similarly, we can compute the conditional distribution
multi_normal_factor.cond_dist(1, f)
(array([-1.13919269, -1.13919269, -1.13919269, -1.13919269, -1.13919269,
0.38187477, 0.38187477, 0.38187477, 0.38187477, 0.38187477]),
array([[0.25, 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ],
[0. , 0.25, 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ],
[0. , 0. , 0.25, 0. , 0. , 0. , 0. , 0. , 0. , 0. ],
[0. , 0. , 0. , 0.25, 0. , 0. , 0. , 0. , 0. , 0. ],
[0. , 0. , 0. , 0. , 0.25, 0. , 0. , 0. , 0. , 0. ],
[0. , 0. , 0. , 0. , 0. , 0.25, 0. , 0. , 0. , 0. ],
[0. , 0. , 0. , 0. , 0. , 0. , 0.25, 0. , 0. , 0. ],
[0. , 0. , 0. , 0. , 0. , 0. , 0. , 0.25, 0. , 0. ],
[0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0.25, 0. ],
[0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0.25]]))
It can be verified that the mean is
Λ @ f
array([-1.13919269, -1.13919269, -1.13919269, -1.13919269, -1.13919269,
0.38187477, 0.38187477, 0.38187477, 0.38187477, 0.38187477])
12.14. PCA and Factor Analysis#
To learn about Principal Components Analysis (PCA), please see this lecture Singular Value Decompositions.
For fun, let’s apply a PCA decomposition
to a covariance matrix
Technically, this means that the PCA model is misspecified. (Can you explain why?)
Nevertheless, this exercise will let us study how well the first two
principal components from a PCA can approximate the conditional
expectations
So we compute the PCA decomposition
where
We have
and
Note that we will arrange the eigenvectors in
𝜆_tilde, P = np.linalg.eigh(Σy)
# arrange the eigenvectors by eigenvalues
ind = sorted(range(N), key=lambda x: 𝜆_tilde[x], reverse=True)
P = P[:, ind]
𝜆_tilde = 𝜆_tilde[ind]
Λ_tilde = np.diag(𝜆_tilde)
print('𝜆_tilde =', 𝜆_tilde)
𝜆_tilde = [5.25 5.25 0.25 0.25 0.25 0.25 0.25 0.25 0.25 0.25]
# verify the orthogonality of eigenvectors
np.abs(P @ P.T - np.eye(N)).max()
4.440892098500626e-16
# verify the eigenvalue decomposition is correct
P @ Λ_tilde @ P.T
array([[1.25, 1. , 1. , 1. , 1. , 0. , 0. , 0. , 0. , 0. ],
[1. , 1.25, 1. , 1. , 1. , 0. , 0. , 0. , 0. , 0. ],
[1. , 1. , 1.25, 1. , 1. , 0. , 0. , 0. , 0. , 0. ],
[1. , 1. , 1. , 1.25, 1. , 0. , 0. , 0. , 0. , 0. ],
[1. , 1. , 1. , 1. , 1.25, 0. , 0. , 0. , 0. , 0. ],
[0. , 0. , 0. , 0. , 0. , 1.25, 1. , 1. , 1. , 1. ],
[0. , 0. , 0. , 0. , 0. , 1. , 1.25, 1. , 1. , 1. ],
[0. , 0. , 0. , 0. , 0. , 1. , 1. , 1.25, 1. , 1. ],
[0. , 0. , 0. , 0. , 0. , 1. , 1. , 1. , 1.25, 1. ],
[0. , 0. , 0. , 0. , 0. , 1. , 1. , 1. , 1. , 1.25]])
ε = P.T @ y
print("ε = ", ε)
ε = [-2.68965844 0.89427629 0.79340353 -0.37962072 -0.82827448 0.65842092
-0.03788382 -0.64394189 0.61617964 0.27127267]
# print the values of the two factors
print('f = ', f)
f = [-1.13919269 0.38187477]
Below we’ll plot several things
the
the
the value of the first factor
the value of the second factor
plt.scatter(range(N), y, label='y')
plt.scatter(range(N), ε, label='$\epsilon$')
plt.hlines(f[0], 0, N//2-1, ls='--', label='$f_{1}$')
plt.hlines(f[1], N//2, N-1, ls='-.', label='$f_{2}$')
plt.legend()
plt.show()
<>:2: SyntaxWarning: invalid escape sequence '\e'
<>:2: SyntaxWarning: invalid escape sequence '\e'
/tmp/ipykernel_8076/2845509298.py:2: SyntaxWarning: invalid escape sequence '\e'
plt.scatter(range(N), ε, label='$\epsilon$')
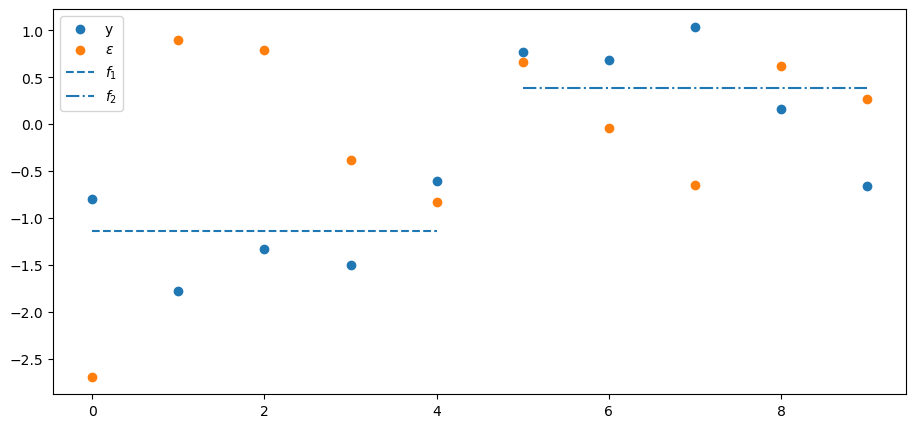
Consequently, the first two
Let’s look at them, after which we’ll look at
ε[:2]
array([-2.68965844, 0.89427629])
# compare with Ef|y
B @ y
array([-1.14557316, 0.38088811])
The fraction of variance in
𝜆_tilde[:2].sum() / 𝜆_tilde.sum()
0.84
Compute
where
y_hat = P[:, :2] @ ε[:2]
In this example, it turns out that the projection
We confirm this in the following plot of
plt.scatter(range(N), Λ @ f, label='$Ey|f$')
plt.scatter(range(N), y_hat, label=r'$\hat{y}$')
plt.hlines(f[0], 0, N//2-1, ls='--', label='$f_{1}$')
plt.hlines(f[1], N//2, N-1, ls='-.', label='$f_{2}$')
Efy = B @ y
plt.hlines(Efy[0], 0, N//2-1, ls='--', color='b', label='$Ef_{1}|y$')
plt.hlines(Efy[1], N//2, N-1, ls='-.', color='b', label='$Ef_{2}|y$')
plt.legend()
plt.show()
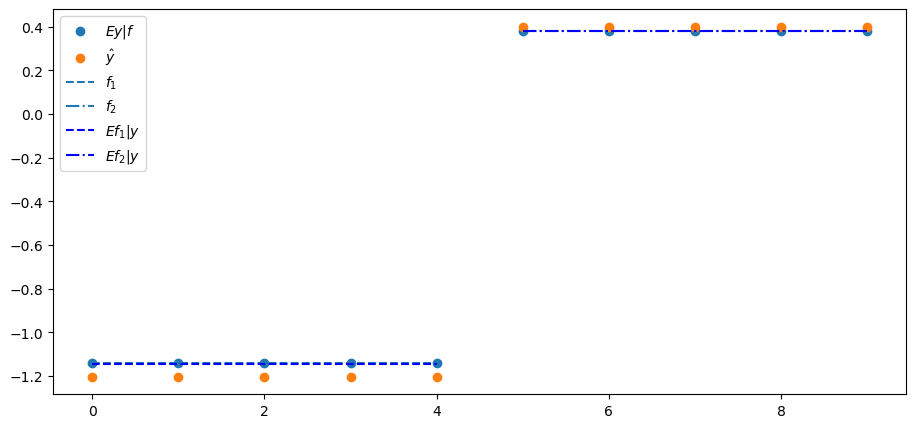
The covariance matrix of
Σεjk = (P.T @ Σy @ P)[:2, :2]
Pjk = P[:, :2]
Σy_hat = Pjk @ Σεjk @ Pjk.T
print('Σy_hat = \n', Σy_hat)
Σy_hat =
[[1.05 1.05 1.05 1.05 1.05 0. 0. 0. 0. 0. ]
[1.05 1.05 1.05 1.05 1.05 0. 0. 0. 0. 0. ]
[1.05 1.05 1.05 1.05 1.05 0. 0. 0. 0. 0. ]
[1.05 1.05 1.05 1.05 1.05 0. 0. 0. 0. 0. ]
[1.05 1.05 1.05 1.05 1.05 0. 0. 0. 0. 0. ]
[0. 0. 0. 0. 0. 1.05 1.05 1.05 1.05 1.05]
[0. 0. 0. 0. 0. 1.05 1.05 1.05 1.05 1.05]
[0. 0. 0. 0. 0. 1.05 1.05 1.05 1.05 1.05]
[0. 0. 0. 0. 0. 1.05 1.05 1.05 1.05 1.05]
[0. 0. 0. 0. 0. 1.05 1.05 1.05 1.05 1.05]]